General options:
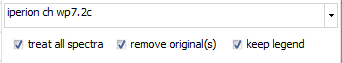
For all functions, you will get a specific window for setting
parameters and options. You will find the top spectrum selector
with all Analyze functions, while the checkboxes only exist with
"Gaussian Deconvolution":
- At the top, there is a dropdown selector containing the current
spectra list. Here you can select the spectrum top be analyzed
(if only one spectrum gets treated). If a preview is available,
the spectrum is selected here.
- if the "treat all spectra" option is checked,
all spectra will get processed and attached as new spectra at
the end of the spectrum list. Otherwise, only the currently selected
spectrum gets processed.
- With the "remove all" option checked, the original
spectra will be removed and only the processed spectra/ spectrum
remain(s).
- The "keep legend" option leaves the original
legend text unchanged. If unchecked, a function-specific text
part will be added to the legend text.
|
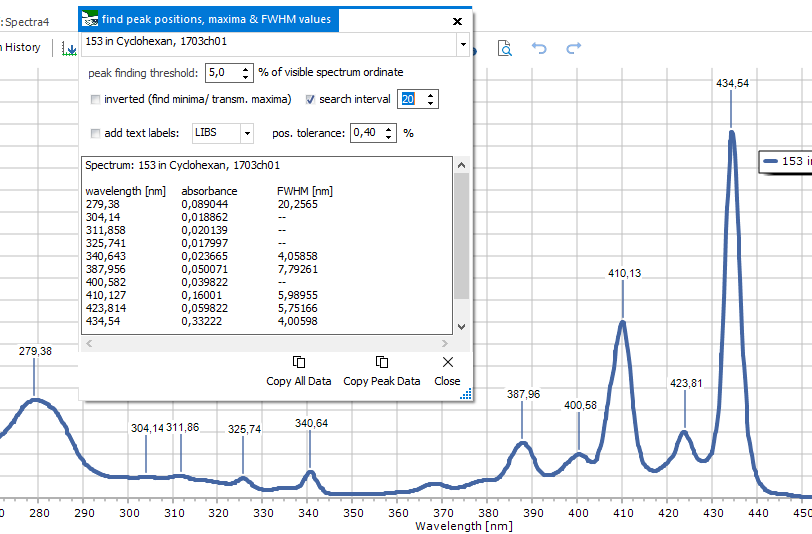 |
Peaks
& FWHM
For the currently
visible x axis range, a list of peak positions together with their
FWHM values is created. A number of parameters can be set:
peak finding
threshold: sets the lower limit of the found peaks, just the
same as it is done with the Peak
labels function.
search interval:
a smaller value finds less prominent peaks, this parameter
corresponds to the prominence parameter from the Peak
labels function.
inverted
(find minima/ transm. maxima): will search for minima
add text
labels: to be used with the special label types, like LIBS,
XRF, Gamma. It adds an additional column to the list with the
respective position labels (atom names for LIBS and XRF, nucleids
for Gamma). On the right, the position tolerance can be set, to
ensure selection of only the proper assignments.
Copy Peak
Data: this button copies the list to the clipboard, to
be pasted for further use into other software.
Copy All
Data: this button copies a concatenated peak list for
all spectra to the clipboard, to be pasted for further use into
other software.
|
| <jump
back to top> |
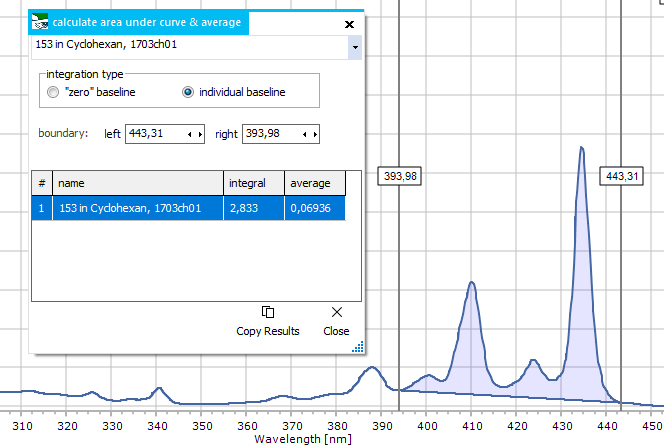
|
Integration
For calculating
the area under curve within a given x axis range. Enter the new
start and end values for the integral range manually, or else
interactively move the two vertical cursors shown together with
the function's window. The results list shows a table of integral
values for all spectra, together with an average value of the
same area (just ignore that if you don't need).
In the plot,
the area under curve gets visualized for the currently selected
spectrum in the upper spectrum selector. The area shape depends
on the chosen integration type:
- "zero"
baseline : uses the x axis as a straight lower boundary for
integration. This only makes sense for spectra with an existing
baseline near zero.
- individual
baseline : uses a straight, but tilted line as lower boundary,
that connects the start and end point of the integral area.
Copy Results:
this button copies the results list to the clipboard, to
be pasted for further use into other software.
|
| <jump
back to top> |
Concentration
(from spectrum)
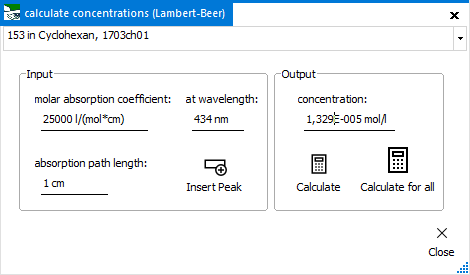
To calculate the
molar concentration for a certain spectrum from its absorbance spectrum.
For this function, the molar absorption coefficient has to be known.
The calculation is done via Lambert-Beer.
All entered
values apply only to the spectrum selected in the upper spectrum
selector. After entry of values for molar absorption coefficient,
wavelength and absorption path length, you can calculate
the concentration with the Calculate button. The
Calculate for all button does the same, but for
all spectra at once (which makes sense for a concentration series
of the same subtance, for example).
Instead of manually
entering the wavelength value, this field can be filled with the
current spectrum's maximum position with the Insert Peak
button.
|
Concentration
(from weighing)
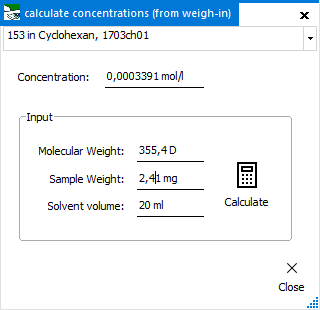
To calculate
the molar concentration for a certain spectrum from its molecular
weight and solution creation. After entering the molecular
weight, sample weight and solvent volume, calculate
the molar concentration with the Calculate button.
The same calculation
is also possible from the Data tab within the Spectra
Properties window, where you can also manually enter the concentration
value.
|
The
calculated concentration value gets saved together with the spectrum
in the *.sgd file format.
HINT: Only with a concentration value assigned, the switching
of y axis type to molar extinction coeefficient or its logarithm
gives a meaningful display! |
| <jump
back to top> |
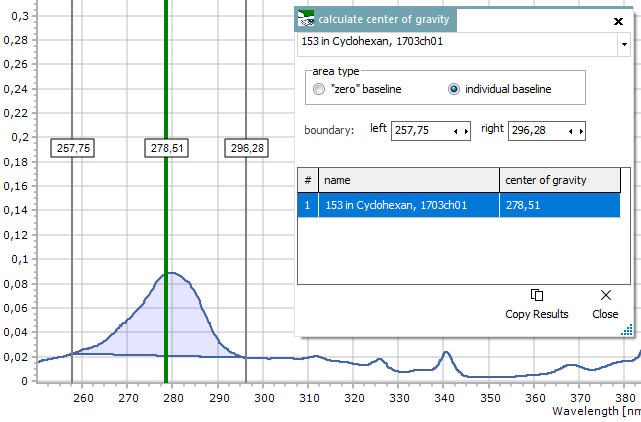 |
Center
of Gravity
For calculating
the center of gravity of a spectrum peak area within a given x
axis range. Enter the new start and end values for the horizontal
range manually, or else interactively move the two vertical cursors
shown together with the function's window. The results list shows
a table of center position values for all spectra.
In the plot,
the area under curve and the center of gravity (green
vertical line) get visualized for the currently selected spectrum
in the upper spectrum selector. The area shape depends on the
chosen area type:
- "zero"
baseline : uses the x axis as a straight lower boundary for
calculation. This only makes sense for spectra with an existing
baseline near zero.
- individual
baseline : uses a straight, but tilted line as lower boundary,
that connects the start and end point of the area.
Copy Results:
this button copies the results list to the clipboard, to
be pasted for further use into other software.
|
| <jump
back to top> |
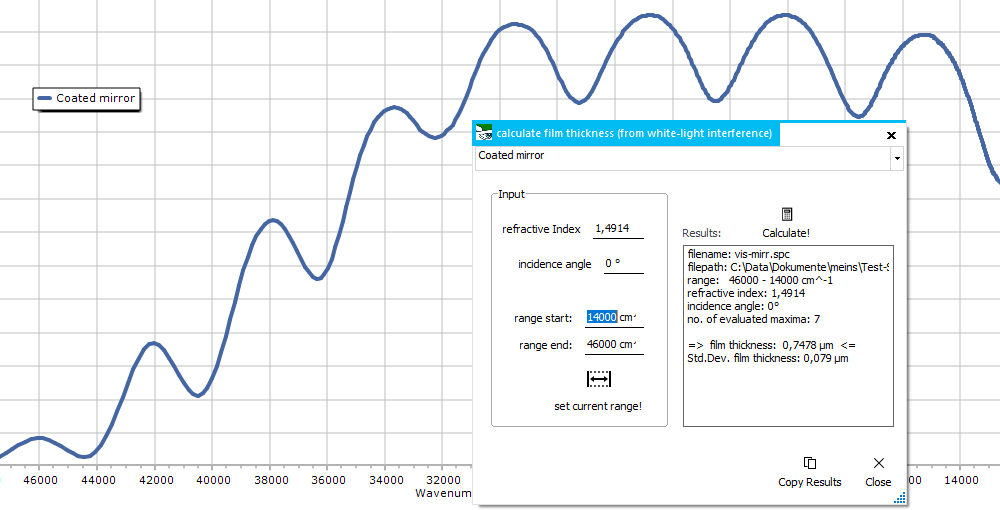 |
Film Thickness
From a white-light
reflectance spectrum showing interference, the thickness of a
single-layer thin film can be calculated.
As input values,
the refractive index of the film material is needed together
with the incident angle of the reflected light (perpendicular
illumination means an angle of 0°) The used wavelength/ wavenumbers
range can be changed manually or by clicking the set current
range button. Clicking the Calculate button
fills the results output window with the calculation results.
The results
window contains the input data, the number of evaluated peaks
and the calculated thickness together with a quality measure (standard
devation).
A StDev value higher than a few percent of the thickness value
indicates an invalid result.
While the calculation
is done on the wavenumbers spectrum, it is no necessary to switch
the display to wavenumbers before calculation.
The
calculation is based on this equation.
It only works for a single-layer thin film! |
 |
|
| <jump
back to top> |
|
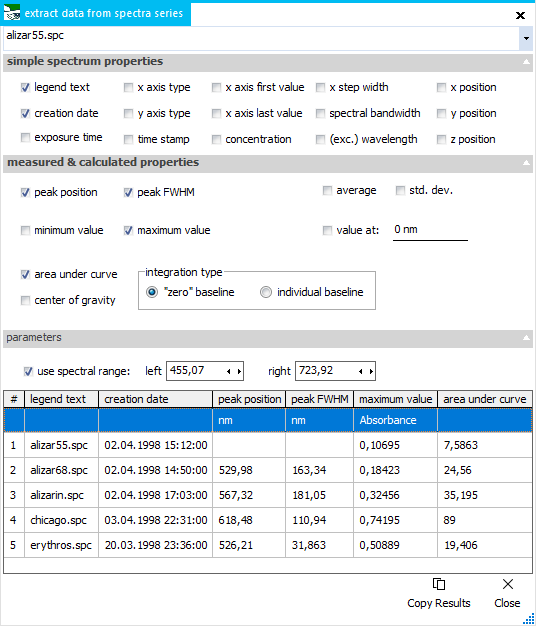
Extract
Data
This function
creates a data table for a loaded spectra series, listing the
values of selected parameters within a given x axis range. After
clicking the Extreact Data button, select the x axis area with
the left mouse (like zooming), then the window is shown. Change
the start and end values for the integral range manually, or else
interactively move the two vertical cursors in the plot. Calculated
properties in the data table will be updated on the fly, when
the spectral range gets changed. As ssecond row, the parameter's
units are shown, if available.
For each selected
parameter, a new column will be created in the data table.
There are parameters
for simple spectrum properties that do not depend on the
spectral range, like legend text (checked per default), creation
date, time stamp, first and alst value and some more.
Then there is
a range of measured and calculated parameters, most of
which are already known from functions elsewhere, like highest
peak position and FWHM, minimum and maximum value, average and
standard deviation, area under curve and center of gravity. Additionally,
the y value at a defined position can be shown, which represents
a spectral cross section of the spectrum series. With "use
spectral range" unchecked, the calculated properties
are taken from the whole spectral range.
Copy Results:
this button copies the data table to the clipboard, to be
pasted for further use into other software.
HINT:
for
a spectrum series with time stamps (like all spectra measured
with Spectragryph from a connected spectrometer), you can easily
create a time series with this function. Just select "time
stamp" as parameter and use this later as x value.
|
| <jump
back to top> |
| |
Create Output
Value
This function
caculates a spectrum property (similar to the Colorize
by property function) and sends it to a physical hardware
as output device. The spectrum property's value range gets mapped
to the value range of the output device, like a voltage or a colour,
or whatever. Selecting another spectrum in the upper spectrum
selector updates the sent output value.
|
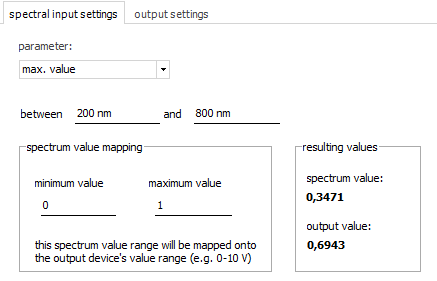
As
parameter, you can select:
- y value (at a defined position)
- minimum value (in a defined range)
- maximum value (in a defined range)
- average of y values (in a defined range)
- standard deviation of y values (in a defined range)
- area under curve (in a defined range)
- peak position (in a defined range)
- center of gravity (in a defined range)
|
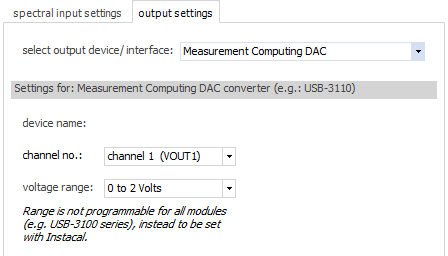
Currently,
there is only one type of output device recognized: the
range of DAC (digital analog converter) devices from Measurement
Computing, like the USB-3110.
You
can define the channel number, as well as the actual voltage
range from the dropdown selectors.
|
HINT:
The
Create output value function is also available
as post-procesing option for live spectra acquisition in the Acquire
ribbon, therefore allowing to continuosly feed a physical
representation of a spectrum property into another system.
|
| |
<jump
back to top> |
 |
Mixture
Analysis
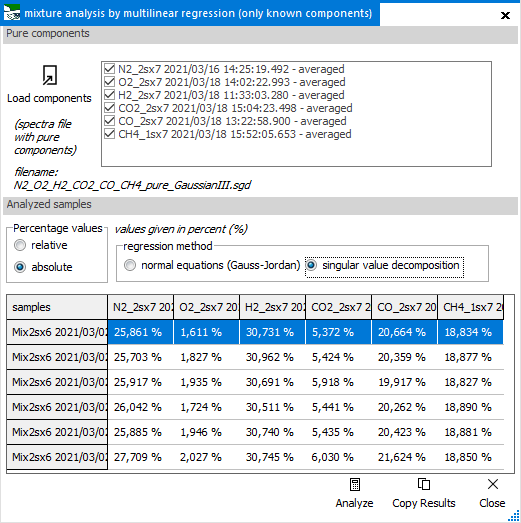
This function
calculates the amounts of known components in a mixture of unknown
composition, based on the given reference spectra for the rpure
components. This done by using a multilinear regression algorithm
(MLR), which can be used with either normal equations, or singular
value decompositon method.
With the Load
components button, the reference spectra of pure compounds
get introduced into the system. You can select or unselect any
of them, to devide which will be used in the analysis. The software
will remember them, so it can ber used repeatedly.
In the data
table below, the analysis results for all loaded spectra are shown
after clicking the Analyze button. The percentage
result can be displayed as relative value, which is the
original result, or else as absolute value, which makes
all entries sum up to 100%.
This is a quite
new feature and needs to be thoroughly validated by yourself with
known data, before going into the unkown. I found that measurement
noise and noisy baselines degrade the results. Therefore I introduced
the Gaussian Deconvolution function (described below), which gives
a noise-free approximation of a spectrum, ideally suited as input
to the MLR algorithm.
|
| <jump
back to top> |
| |
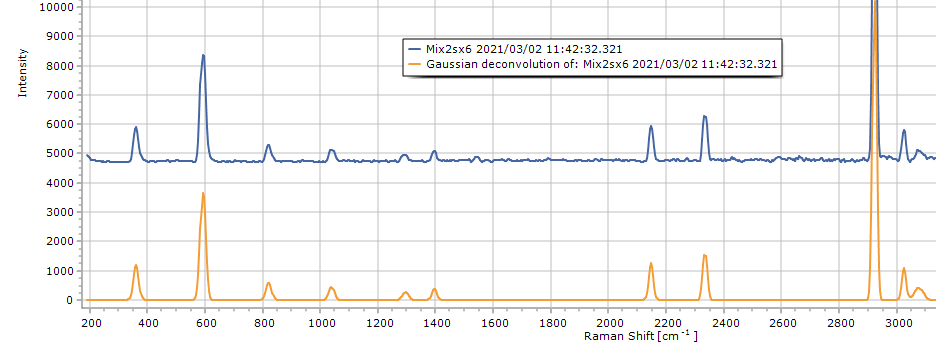
Gaussian Deconvolution
This function
creates a noise-free spectrum approximation based on Gaussian
peaks replacing actual spectrum peaks. This is not (yet) an iterative,
fitting algorithm, so "deconvolution" is probably no
the right wording. Instead it simply looks for spectrum peaks
and creates a Gaussian curve instead, based on peak height and
width. It works pretty well on Raman spectra with not too many/
not overlapping peaks and can be used as input for the mixture
analysis function (described above). Two alternative ways for
peak finding area available:
- percentage of max-min: takes a relative lower threshold
and combines that with a prominence values (as known from the
Peak Labels
function)
- baseline noise level: caculates the lower threshold from
a multiple of the baseline noise in a user-defined area
 |
| |
<jump
back to top> |
| |
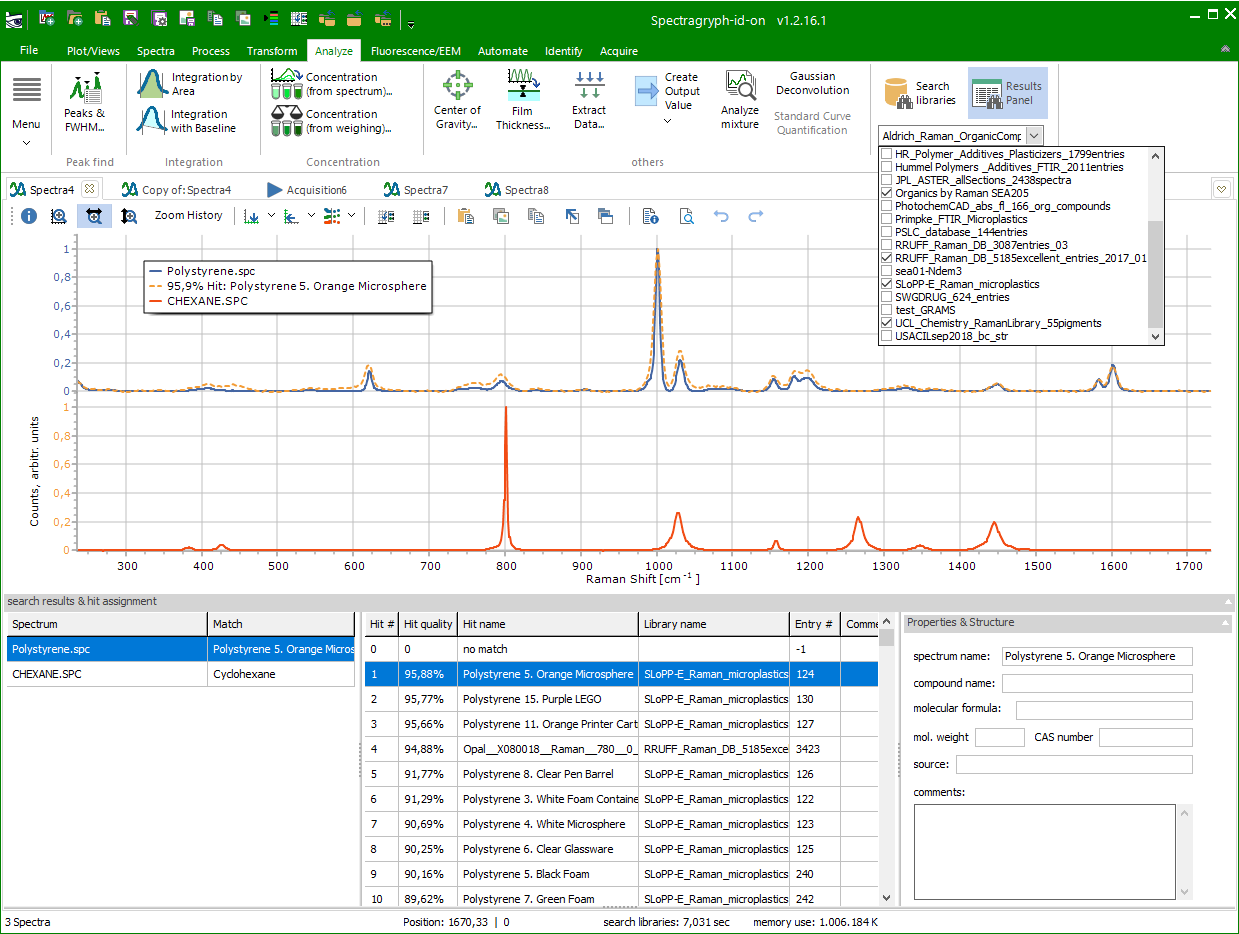
Identification
(through spectral library search)
This area allows
multi-library search for all loaded spectra in an ordinary spectra
tab, with display of found hit names and values and library spectra
within the plot.
First, select
the libraries to be searched in the dropdown library selector,
which shows the names of all libraries in the libraries folder
(usually: C:\Users\Public\Documents\Spectragryph\Libraries\,
to be defined in the Identification options  ).
In options, you can also define the match threshold, and
whether to display found reference spectra in the plot
and if they should be normalized to the sample spectra. ).
In options, you can also define the match threshold, and
whether to display found reference spectra in the plot
and if they should be normalized to the sample spectra.
Then, click the Search Libraries button for actual
searching, a progress indicator in the lower status bar shows
whats going on. When finished, the Results Panel is filed with
the search results, and the best hit for the first spectrum is
shown in the spectrum plot.
The lower results
panel shows this interactive content:
- the spectra list (at left) with their best hit (or "no
match") if no hit above the match threshold was found
- the hit list (in the middle) showing the best
20 hits for the selected spectrum, with their hit quality value,
spectrum name, library name, and so on
- the properties & structure information (at left)
for a selected hit, as found in the spectral library
Scrolling (left
mouse click, mouse wheel or keys) through the hit list will update
the displayed reference spectrum above.
Clicking on a spectrum or its legend text in the plot, or selcting
it in the spectrum list (at left in results panel), will update
the plot to show the assigned hit as reference spectrum, with
the hit quality as part of the legend text.
HINT:
the same multi-library search and display of results is also available
as post-processing function while acquiring spectra in the Acquire
ribbon. Therefore you can have live sample identification
during continuous measurement.
|
| |
<jump
back to top> |
|
![back to SpectraGryph main page [SpectraGryph]](gryphon_white_green_96.png)
![back to SpectraGryph main page [SpectraGryph]](gryphon_white_green_96.png)